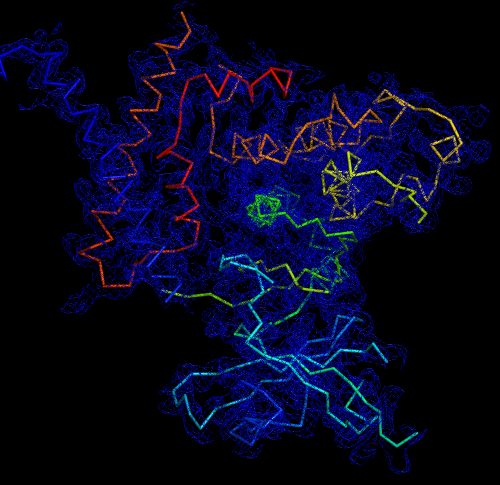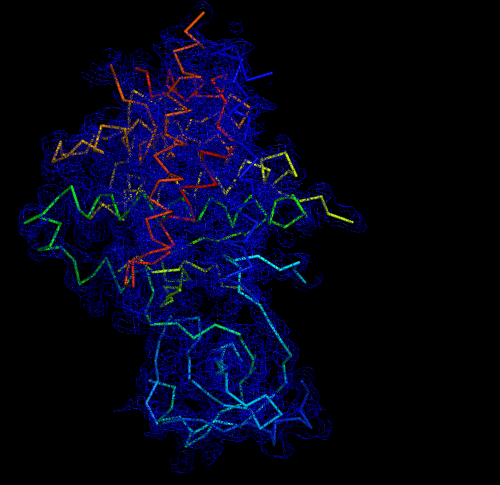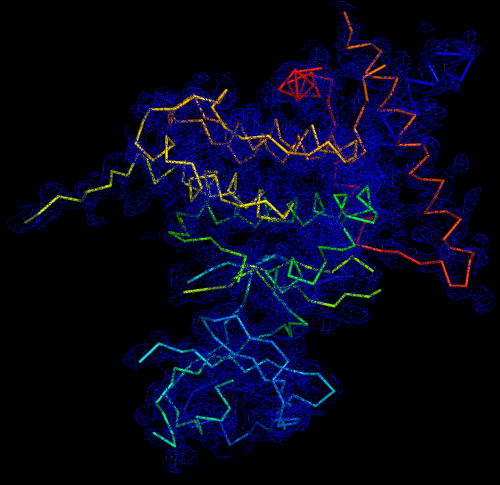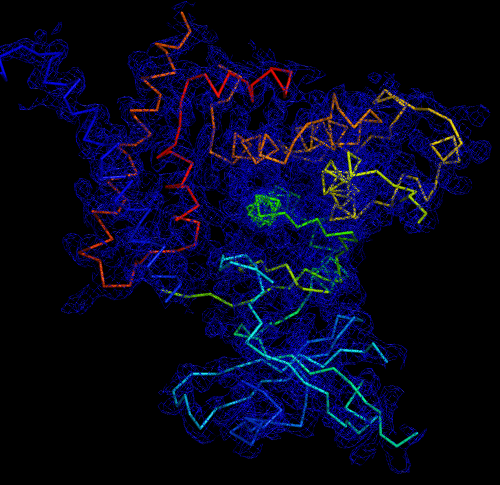
Melbourne researchers have used the Australian Synchrotron to produce the first three-dimensional structure of a molecular scaffold, known to play a critical role in the development and spread of aggressive breast, colon and pancreatic cancer.
Armed with the structure, the research team is looking at ways of targeting parts of the scaffold molecule critical for its function. They hope the research will lead to novel strategies to target cancer.
The research was the result of a long-standing collaboration between Walter and Eliza Hall Institute (WEHI) researchers Dr Onisha Patel and Dr Isabelle Lucet and Monash University Biomedical Research Institute researcher Professor Roger Daly.
Dr Santosh Panjikar, a macromolecular crystallographer at the Australian Synchrotron and Dr Michael Griffin from Bio21 Institute at the University of Melbourne made important contributions to the study, which was published in the journal Nature Communications.
Lucet said SgK223 is a member of a family of proteins called pseudokinases and had been classified for a long time as a ‘dead enzyme’.
“SgK223 doesn’t have the measurable activity that we see with other types of enzymes, and this meant it was largely ignored. However in the past decade, we’ve come to understand that this ‘dead enzyme’ plays an active and important role in cell signalling,” Lucet said.
MX2 beamline used to determine crystal structure
Panjikar explained that measurements on the macromolecular crystallography beamline, MX2 and small angle X-ray scattering on the SAXS/WAXS beamline were used to determine the crystal structure of SgK223 and oligomeric state of SgK223 in solution respectively.
“There were many challenges working on this protein,” said Panjikar.
Initial experiments on crystals of SgK223 were not successful because the protein is highly sensitive to various heavy atom solutions, and to X-ray radiation and deteriorates very quickly.
“Protein crystals are normally sensitive to radiation but the selenomethionine protein crystals were more sensitive,” said Panjikar.
“You want to collect your data in a way that doesn’t damage the crystal but retains the anomalous signal,” said Panjikar.
"We designed a diffraction strategy for the sample, in which we used a small sized beam 20 microns across and clad at several places going from one position to the next on the rod-shaped crystals.”
The researchers collected multiple data sets at different X-ray energies around selenium edge for multiple wavelength anomalous dispersion (MAD), a specialised technique that allows them to use ‘tunable energy’.
“Selenium has an absorption edge at particular energy where it absorbs more X-rays. We went on to collect X-ray data from Sgk223 crystal at the higher energy side of that edge and also at below the edge,” explained Panjikar.
They were able to solve the crystal structure using a software pipeline, Auto Rickshaw, developed by Panjikar.
“Where you have multiple data sets at different energies, you need to check which data set will actually work. In this case, with Auto Rickshaw we found one combination of the data sets that worked very well,” said Panjikar.
The data was used to get the preliminary phases and electron density map, which enabled the researchers to build the 3D model.
“When you determine the crystal structure of the protein you know what the molecule looks like but it also confirms if the molecule is a monomer or dimer,” said Panjikar.
SAX confirms crystal structure
“We could see that SgK223 was a dimer, but needed supporting confirmation that what we saw in the crystal structure was the same in solution. “
Validation of the dimer was achieved using small angle X-ray scattering of the native protein.
Other biochemical techniques carried out at WEHI, Monash and the University of Melbourne were used in the study.
“The world-class facilities at the Australian Synchrotron in Melbourne were instrumental in the discovery,” co-author Lucet said.
“It is the only facility in the Southern Hemisphere that has the specialised technology required to provide us with detailed knowledge essential for seeing molecules at an atomic level. This is essential if we wish to discover and develop drugs that target and interfere with molecules that drive cancer and other diseases.”
Media release
Read more about the research on the Walter and Eliza Hall Institute of Medical Research.
DOI: 10.1038/s41467-017-01279-9
